Get your large SQL data in ff swiftly
The ff package is great when you are working with large data in R. Data in corporate environments are usually not that large that a Hadoop system is needed to handle it but the data are mostly large enough to make R choke on it's RAM.
The ff package is great for this type of data. It can handle 2.14 billion elements per atomic (so 2.14 billion records). This is mostly more than enough in standard business settings nowadays.
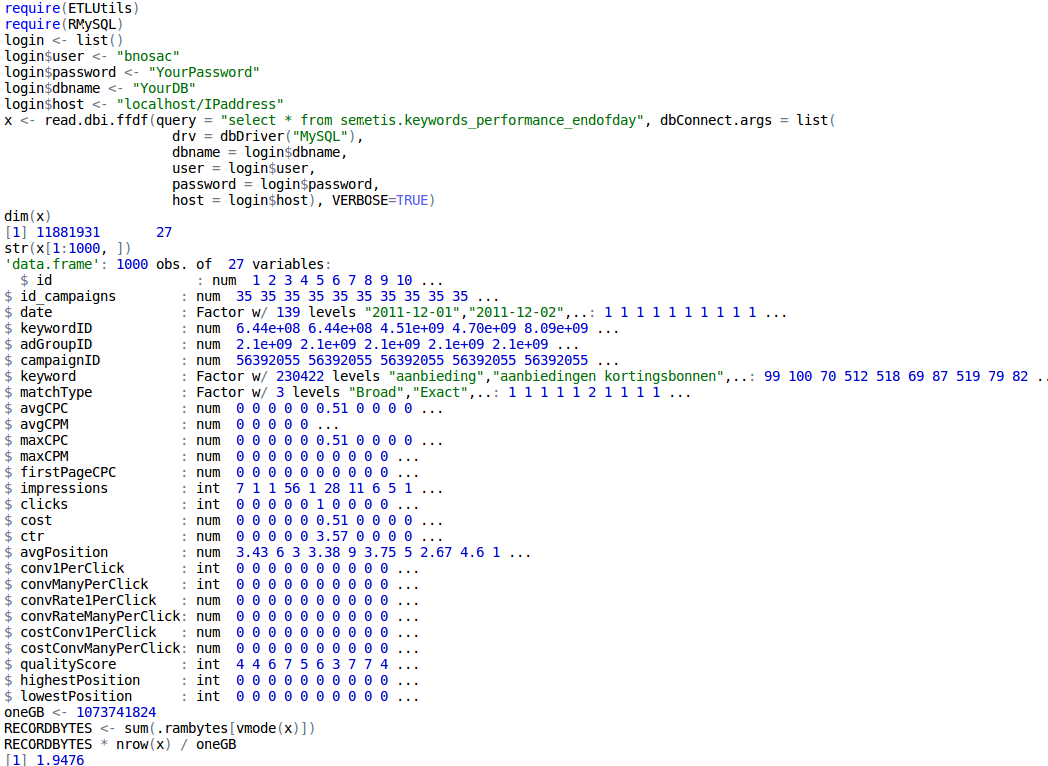
For a while getting the data in R was quite cumbersome. The ff package now provides already nice interfaces to csv and flat files in the read.csv.ffdf and read.table.ffdf functions. To extend this towards standard SQL databases, the ETLUtils package has been put at CRAN. It contains a function called read.dbi.ffdf which can be used to extract easily data from any sql database through DBI directly into an ffdf in R.
Below, we show an example of importing a table stored at a MySQL database with 11.8 million records directly in an ffdf without having RAM issues.